
Combining programming and biology
Unlike people who might see problem-solving as a frustrating process, Dr. Gregory Schwartz considers solving problems to be a fun challenge.
Gregory's odyssey into the world of problem-solving began early. He was captivated by computer games, seeing them as a playground for honing his analytical abilities, which ultimately led to a long-lasting interest in programming and computer science.
Shortly after, in high school he was introduced to a new passion: biology. When Gregory needed to choose between which of these two passions he wanted to pursue as a career, his great uncle, a podiatrist who applied mathematics to his own research, pointed Gregory to the burgeoning field of bioinformatics.
“That's how I learned that you could combine computational analysis with biological questions,” says Gregory. “I always knew I wanted this as my career. I just didn't know it existed and I didn't know what it was called.”
Gregory received a double major in mathematics and biology during his undergraduate studies in university and later embarked on bioinformatics projects during his Masters and PhD degrees at Drexel University in Philadelphia, USA.
During his PhD, Gregory delved deep into the world of immune-cell receptors. Adaptive immune responses from cells, such as B and T lymphocytes, can be triggered when their cell surface receptors recognize and bind to materials from invading pathogens. Therefore, the body has a diverse genetic repertoire of lymphocyte receptors to fight against a variety of pathogens. Gregory’s research focused on quantifying the diversity of these receptors to understand how receptor variability affects an individual’s response to specific pathogens.
It was during his PhD that Gregory started to develop computational tools to quantify the diversity of biological systems. He looked at the genes that encode lymphocyte receptors, clustered different regions by their structural similarity and came up with a mathematical measure to quantify how similar genetic regions are to one another.
This mathematical measurement, simply called “clumpiness” by Gregory, has been constantly applied to his later work.
Mapping the social network of cancer cell communication
Gregory’s research interests extended beyond immune-cell receptors and branched into the realm of cancer biology during his postdoctoral studies at the University of Pennsylvania under the supervision of Dr. Robert Faryabi, an expert in epigenetic regulation of cancer-related gene expression.
To quantify different kinds of diversity within cancer cells, Gregory developed four computational tools: HeatITup, a tool that looks for a particular kind of mutation called internal tandem duplication mutation in cancer cells; inteGREAT, an algorithm that integrates different data modalities like transcriptome and proteome data to identify new cancer biomarkers; TooManyCells, a platform to quantify the heterogeneity and diversity among single cells using gene expression data; and TooManyPeaks, which groups cells by chromatin accessibility similarity.
The tool that Gregory is most proud of is TooManyCells. It uses data generated from single cell RNA sequencing to group cells based on their gene expression profiles. So far, it has been used to show how T-cell acute lymphoblastic leukemia (T-ALL), a type of aggressive blood cancer, develops resistance to therapy.
Currently, gamma secretase inhibitor (GSI) is being tested a potential treatment for T-ALL; however, studies have shown that some T-ALL cells can develop resistance to the drug. To better understand this, Gregory used TooManyCells to compare the gene expression profiles between untreated T-ALL cells and T-ALL cells that were treated with GSI and developed resistance. He found that the resistant, treated cancer cells had similar gene expression profiles to untreated cancer cells.
“This helps us to understand how we can overcome therapeutic resistance,” says Gregory. “There is a subgroup of cancer cells that may tolerate the therapy, and a selection process may happen when GSI treatment is applied. If we can stop the resistant subgroup from forming, then we can improve the effectiveness of the therapy.” The study was published on Nature Methods in 2020.
After completing his postdoctoral studies in 2021, Gregory joined the Princess Margaret to establish his own lab where he looks for and analyzes different cancer-cell populations within the same tumour. He aims to expand methods for understanding the differences between cancer cells, by exploring a wider array of datasets, including temporal and spatial information about tumours.
In previous projects where he sought to identify the most commonly mutated genes in T-ALL and colorectal cancer, he noticed that the top mutated genes code for crucial receptors that play a role in cell–cell communication. “No matter which cancer type we look at, some of the most highly expressed genes are those involved in cell–cell communication. This prompted us to develop a way to quantify how cells talk to each other and to surrounding cells, or stroma within a tumour, and determine whether those interactions correlate with disease progression and therapy resistance.”
To figure out when and where cancer cells communicate, Gregory is developing tools to visualize spatial relationships in various tumour regions. In a collaboration with Dr. Faiyaz Notta’s lab at the Princess Margaret, the two teams are trying to detect cancer cell-communication signals in pancreatic cancer samples.
A postdoctoral researcher in Gregory’s lab, Dr. Fatema Zohora, has recently integrated spatial and molecular data to develop a visualization tool to map cancer-cell communication in a pancreatic tumour sample (illustrated below).
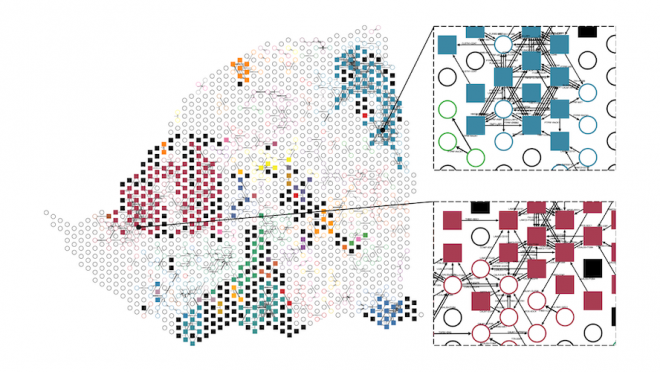
An example of pancreatic ductal adenocarcinoma tissue. Filled squares are tumour cells, open circles are stroma (normal) cells and the black lines with arrows are communication indicators inferred by the visualization tool.
“We can see that there are a variety of different signals passing across tumour and stroma regions. Importantly, each tumour region can express distinct communication pathways, suggesting that we need to use high-resolution techniques to fully understand how tumours survive treatment. We can then use our tools to identify specific survival mechanisms of a tumour within a patient to help guide therapy.”
Gregory and his team are studying how cancer cells change their communication patterns when they become resistant to treatment. By deciphering these communication networks, they hope to develop personalized treatment strategies for cancer patients. Ultimately, their goal is to create a precise roadmap for treating different types of cancer by targeting the specific cell signals that drive them, bringing us one step closer to more effective and tailored cancer therapies. And it is worth noting that they are doing all of this in an open-source, collaborative way, contributing to the broader scientific community's understanding of cancer biology.
Establishing a lab as an early career investigator
Gregory embarked on an exciting journey when he relocated to Toronto to start a career as a new independent investigator.
His move to Toronto was marked by a profound appreciation for the city and its vibrant academic environment. “The University of Toronto and the research hospitals within the University Health Network are all located in a ten-minute walking distance. You barely need to move to immerse yourself in a variety of talks and stimulating academic discussions,” Gregory expressed with excitement.
Gregory's transition into his role as an independent researcher is remarkable. While searching and applying for jobs near the end of his postdoctoral work, Gregory attended many interviews at different institutions and PM was a clear standout.
“I was very blown away by the interview process here at the Princess Margaret,” Gregory recalls. “Everyone treated each other like family. They were deeply engaged and interested in my research, proposing new projects and making suggestions even at the interview stage. It was such a fantastic experience, and it was immediately clear in my mind that this would be the ideal place to build my research career.”
As a new investigator, Gregory found himself in the unique position of supporting and mentoring others, which he finds immensely rewarding. Watching his team members learn and grow is a source of pride and joy.
However, he also acknowledges that the transition came with challenges. Starting from scratch meant acquiring new skill sets, such as managing research teams, navigating the hiring process and honing grant writing techniques. Despite the initial uncertainties, Gregory leaned on the support of mentors, colleagues, and fellow scientists who were also starting their own journeys as new independent researchers. Together, they formed a tight-knit community, reinforcing the idea that research is a collaborative effort that extends beyond individual projects.
“It was scary coming in with the need to establish everything from scratch,” says Gregory. “Thankfully, my mentors and colleagues here have been extremely helpful. I have never felt isolated in this process.”
Meet PMResearch is a monthly column that features Princess Margaret researchers. It showcases the research of world class scientists, as well as their passions and interests in career and life—from hobbies and avocations to career trajectories and life philosophies. The researchers that we select are relevant to advocacy/awareness initiatives, or have recently received awards or published papers. We are also showcasing the diversity of our staff in keeping with UHN themes and priorities.

