By: S. Amanda Ali, UHN Trainee and ORT Times Writer
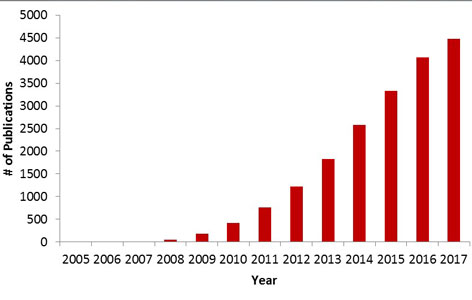
Have you ever considered having your genome sequenced? In theory, it could provide you with information about your genetic predispositions for a variety of factors, including disease. It would only cost around $1,000 USD and could be completed in less than a day. The technology is known as next generation sequencing (NGS), a massively parallel method for determining the exact order (or sequence) of nucleotides in your DNA. Sequencing can also be done for RNA, protein, and carbohydrates, as it essentially gives you primary structures in the form of a sequence of monomeric subunits. Over the past 10 years, the use and reporting of NGS has drastically increased (Figure 1).
So why is NGS so popular and what does it mean for society? Before answering this question, let’s take a step back: if this is next generation sequencing, what came before? Sequencing was first performed using a method known as Sanger sequencing, which was developed in 1977. This method uses modified nucleotides that are incorporated during DNA replication to terminate the process and to create short sequences of DNA fragments that are aligned and compiled to create a single sequence. This sequencing method has significant limitations in terms of yield, cost and effort required. With NGS and its massively parallel method, the yield is significantly higher (more bases sequenced in less time), and the cost and hands-on time are significantly lower.
NGS, which is also known as high-throughput sequencing, has been achieved using different technologies, including Illumina/Solexa sequencing, Roche 454 sequencing, Ion torrent sequencing and SOLiD sequencing. Most common perhaps is Illumina, which uses ‘sequencing by synthesis’ technology that is quite similar in principle to Sanger sequencing. Again, modified nucleotides are successively added but rather than permanently terminating DNA replication, these nucleotides can be imaged one at a time (as they are fluorescently labeled), so that replication can continue and the next nucleotide can be added and imaged. This allows multiple DNA fragments to be sequenced simultaneously and leads us back to the major advantages of NGS for whole genome sequencing – higher output in less time and for lower cost.
This improvement in technology explains the recent surge in NGS (Figure 1). Now we can know the sequence of our DNA for about the same price as an all-inclusive vacation to the Caribbean and in the amount of time it might take us to pack our bags. But what information are we really gathering by knowing the sequence of our DNA? Will it tell you whether you are more susceptible than others to a rare disease that is endemic to your vacation destination? Or how fast you’re going to burn while tanning at the beach?
Maybe someday, just not today.
While it is critically important that we understand the value of NGS for improving our understanding of health and disease, it is equally important to maintain realistic expectations about what information can actually be revealed through NGS. This can be likened to learning a new language. Being able to read the letters does not mean you will understand the words or sentences that are encoded by those letters. Progress is being made daily in research settings around the world to apply experimental and computational approaches to understand the rich information that is encoded in our DNA (e.g. we now know that mutations to the BRCA1 and BRCA2 genes impact breast cancer susceptibility). Continued efforts in laboratories will improve our capacity to not only read, but also interpret and understand our complex genetic code.

DNA Double Stranded Helix. Image Courtesy of: https://www.maxpixel.net/Dna-Network-Research-Chemistry-Medical-Biology-3539309.




